Research Topics
Polar Pelagic Organisms
We are collaborating with the Alfred Wegener Institute and the Carl Von Ossietzky University Oldenburg to create and annotate de novo transcriptome assemblies of krill species.
We have recently released KrillDB2 (website, paper), an updated version of the transcriptome of the Antarctic krill Euphausia superba. We have collected all publicly available sequencing data on this species and we have performed a meta‑assembly, an hybrid computational approach which aims at combining the results of multiple transcriptome reconstruction tools (five, in our study). The resulting transcript collection significantly expands our understanding of the complex krill life cycle. Of particular relevance, we have identified six novel putative opsins including a short-wavelength sensitive variant (SWS/UV) which was never reported before. We also provided the first evidence of microRNAs in krill and their role in defining the adaptive capabilities of this species.
The focus of our investigation is now broadening to include more krill species from other latitudes. We are working on a comparative study to highlight the effects of the adaptation to different climate conditions.
Visualization of Biological Networks
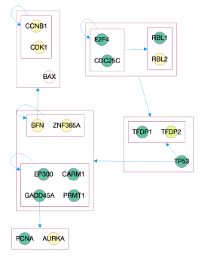
Understanding complicated networks of biomolecular entities and interactions is essential to solving contemporary problems in biology, especially in computational domains such as Systems Biology. Complexity arises due to the sheer size of networks and to the multitude of entities being represented, that range from individual reactions to entire pathways. It is becoming increasingly difficult to interpret the information encoded in networks and to cross‑link their contents across different data sources.
We are exploring dynamic layout algorithms to reduce the clutter present in contemporary visualization formats. We apply graph compression techniques to condense large graphs into a reduced set of visual symbols. The user is then able to selectively increase the level‑of‑detail of the representation around specific areas, following the needs of his analysis.
A fundamental aspect of this work involves the accessibility of the software we build. We decided to rely on web technologies and standards; our viewer is designed to be embeddable in any webpage. As a showcase of its features, we are extending the GraphiteWeb analysis service (website, paper) with new graphical capabilities.
Spatial Transcriptomics
Spatial transcriptomics promises unprecedented insights into cell biology thanks to the possibility of preserving the spatial context of gene expression measurements. While numerous experimental protocols have emerged recently, the landscape of available methods for data analysis remains severely fragmented.
We have started an effort to improve access to different computational methods through a uniform interface, the Bioconductor platform. Our first result is SpatialDE, a wrapper package for using the similarly named statistical test implemented in Python.
In collaboration with Davide Risso's group, we are also evaluating the performances of different methods that have been proposed for the analysis of spatial gene expression.